Abstract
It
has long been known that for certain narrowly defined problems, special-purpose
processors can achieve significant cost/performance gains over general purpose
machines. We describe one such processor
here: a custom accelerator for gene sequence analysis,.
The
use of rapid prototyping technology was crucial in our efforts to produce a
working system in a short amount of time.
Field programmable gate arrays and in-house PCB prototyping were of
special importance. We discuss how these
technologies were used in our system, and offer price-performance comparisons
of our processor with other machines. The processor is completely functional,
and yields a 15x performance improvement
over an unassisted host.
1.0 Introduction
Recent advances in molecular
biology have made computational accelerators for gene sequence comparison of
growing importance. Biologists will soon
be capable of sequencing the entire human genome, producing billions of bytes
of information and demanding large amounts of computing power.
Unfortunately,
computer architects have not matched this demand for increased computing power
with improved architectures. Biologists
must content themselves with personal computers, which offer low cost and low
performance, or supercomputers, which perform well but are extremely
expensive. There is little middle
ground.
To address
this problem, we have constructed a machine that costs as much as a personal
computer but offers an order of magnitude improvement in performance. This machine, the Gene Sequence Processor,
attaches to a host computer and can compare gene sequences 15x faster than the
host alone, for a cost of around $2,000.
Our machine is also "alignment-preserving"; it permits the
user to see how sequences are related to one another in addition to computing a
numerical similarity metric. We will
explain this in more detail shortly.
2.0 Previous Work
The past few years have seen a
dramatic increase in the efforts of biologists to use computers for gene
sequencing. The Journal of Nucleic Acids
Research produces a special issue every two years devoted exclusively to
computer related research. There is now
a special journal for the applications of computers in biology [1], as well as
books devoted to computers and gene sequence analysis [2].
Virtually
all this work is concerned with software and algorithms. Research in the area of architecture has
been less active, probably due to the relative inaccessibility of architectural
ideas to biologists. One of the first
attempts was made by Collins and Coulson [3], who suggest using the DAP SIMD
machine, for sequence processing.
Mukherjee [4] discusses efficient
hardware algorithms for finding the longest common subsequence of two strings,
although no performance results are reported.
Many supercomputers have received careful attention from researchers,
including the Hitachi S810-20 [5], and the iPSC/1 and the CM-1 [6]. Lopresti discusses the performance of the
Multiflow Trace, Convex C1, Cray-2, and CM-2 on gene sequence comparison
problems in [7]. We shall have more to
say about these results in later sections.
Perhaps the
most successful effort in the area of special purpose processing is the Splash
highly parallel programmable logic array, developed by Lopresti and colleagues
at the Supercomputing Research Center [8].
This machine evolved out of the Princeton Nucleic Acid Comparator, or
P-NAC, developed by Lopresti and Lipton [9].
Both of these machines use a systolic array architecture to calculate
the diagonal values of the dynamic programming array in parallel. Both Splash and P-NAC were prototyped
successfully, although neither machine is alignment-preserving. We will compare their price and performance
with the Gene Sequence Processor shortly.
3.0 The Gene Sequence Processor
The Gene Sequence Processor was
designed and built at the Thayer Rapid Prototyping Facility, a laboratory dedicated to the rapid
production and evaluation of digital systems.
This laboratory has been involved in several successful experiments in
rapid digital system design. In addition
to the Gene Sequence Processor, other projects include a completely functional
DLX microprocessor, an FHT transform engine, and a real-time data processor for
rocket telemetry. For further
information on the RPF and these projects, the reader is referred to [10],
[11], and [12].
The GSP consists of five basic
subsystems: DRAM, SRAM, a counter/comparator, the host interface, and the Gene
ALU. It implements a dynamic programming
algorithm to compute the minimum cost sequence of insertions, deletions, and
substitutions necessary to transform one nucleotide sequence into another,
based on a set of cost functions supplied by the user. The total cost of this sequence of operations
is referred to as the edit distance between A and B. For more information on sequence comparison
algorithms, the reader is referred to [13] and [14].
The GSP
contains 8 megabytes of DRAM, organized as a 4M x 16-bit words, limiting the
product of the lengths of the sequences to be compared to 222.
The DRAM holds the dynamic programming
array. Each byte of a DRAM word is
broken into 6 bits of data and a 2 bit field called a back pointer. Back pointers
are used to reconstruct the minimum cost sequence of operations necessary to transform one sequence into
another. For more information on back
pointers the reader is referred to [6].
Back pointer
information was made identical in each byte to simplify the design of the
processor, leaving 12 bits of data in each array entry. Thus the maximum edit distance the GSP can
compute is 212 = 4096.
Currently, cost function values are constrained to be integers. This has not proven to be a limiting factor
in our experiments.
The
sequences to be compared, cost function values, and sequence lengths are
downloaded into a 4k x 8 SRAM. The
counter/comparator is used to generate
DRAM addresses for the dynamic programming calculation, and to halt the
computation when the last entry in the dynamic programming array has been
calculated (the location of the last entry varies with the size of the
sequences to be compared). The processor
interface is responsible for all interaction with the host.
The Gene ALU
is an optimized computational element tailored to the basic operations of the
comparison algorithm. The relevant cost
coefficients and DP array values are added in parallel, and sent to a 3-way
comparator that determines the minimum value.
This value is then written back into the DRAM dynamic programming array.
The Gene ALU
is bit-sliced into upper and lower bytes, with each slice implemented on a field programmable gate array, or FPGA. FPGA's have proven extremely valuable in our
design; we anticipate these devices to play a major role in microsystem
prototyping over the next few years.
Accordingly, we discuss their use in the next section. Readers unfamiliar with FPGAs are referred to
[15] and [16].
4.0 FPGA Usage in the Gene Sequence Processor
The
Gene Sequence Processor uses 5 Actel FPGA's: 3 ACT1020's, 1 68-pin ACT1010, and
1 44-pin ACT1010. (Actel's
antifuse-based architecture is described by El Gamal and El-Ayat in [16] and
[17]). FPGA utilization in the GSP is as
follows:
Gene ALU: 2 ACT1020's, used for
adder (3), latches, and a 3-way comparator.
1 FPGA for each byte.
Counter/Comparator: ACT1020 (1),
used for 16-bit counter (2), 16-bit comparator (2), register.
SRAM interface: ACT1010-68, SRAM
selector & latch, 16-bit 2-1 mux & latch.
DRAM RAS/CAS mux: ACT1010-48,
10-bit 2-1 mux
We observed
the following benefits from using FPGAs in the GSP:
1)
Reduced board area. Our design was constrained at the outset to
fit on two boards of fixed size.
Implementing the random logic of the Gene ALU alone, using discrete
components, would have exceeded these constraints.
2) Reduced chip count. The 5 FPGAs listed above replaced
approximately 50 TTL components. This
reduced design complexity and bringup time (see #5).
3)
Reduced power consumption. The
FPGAs consume substantially less power than the components they replace.
4)
Increased flexibility in design partitioning. FPGAs in a design can be viewed as boxes
whose contents can change as the design evolves. It was common in the design of the GSP to
migrate various pieces of the design on and off FPGAs as the design changed.
5)
Reduced bringup time. All our
FPGAs worked correctly the 1st time they were programmed. The reliability of the devices eliminates
many sources of errors during system bringup.
The use of FPGAs to replace large numbers of discrete components means
fewer connections, fewer nets, and easier debugging.
For a more
detailed discussion of the advantages and disadvantages of using FPGAs in
digital designs, the reader is referred to [10], [12], and [18].
5.0 Rapid PCB Prototyping Technology
The RPF is
perhaps unique as an academic laboratory in that it has the capability of
producing printed circuit boards on site, in the same room in which systems are
designed. The RPF employs a PCB
prototyping system developed by Direct Imaging Incorporated, in which a
resistive ink is sprayed on copper sheets and then etched with sodium
persulfate. The ink is then scrubbed
off, the sheets tin plated and automatically drilled, and then assembled into a
finished prototype.
The GSP
consists of two boards, both made completely in-house at the RPF.
No external fabrication was required.
Netlists of the simulated design are input to a PCB layout program,
which produces a Gerber file. This file
is then input to the PCB prototyping system, running on an IBM PC. The system uses a special printer to spray
ink onto a copper sheet. This sheet is
then rinsed with sodium persulfate, etching the copper away where the ink does
not protect it. After the etching
process, the ink is scrubbed away with steel wool, and the sheets immersed in a
tin solution to plate the traces.
Once the sheets
are dry, they are lined up and drilled.
Drilling takes place on an NC drilling machine supplied with the PCB
prototyper. The drilling process is
completely automatic, under control of the PC.
User intervention is required only to mount new sheets and change drill
bits. When the sheets are drilled, they
are lined up using an optical punch and laminated. Pins are inserted into the appropriate holes,
sockets are soldered onto the board, and the system is ready for IC's and
testing.
The
existence of in-house PCB prototyping technology makes rapid prototyping
feasible for research facilities that do not have access to on-site VLSI
fabrication. The turnaround time for
boards becomes hours instead of weeks, a considerable improvement over using
remote facilities for system development.
6.0 Performance Results
The current implementation of GSP
consists of two boards that plug in to the NuBus slots of a Mac II f/x personal
computer. The performance of these
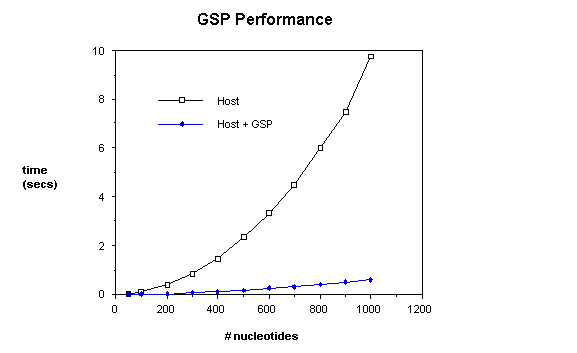
boards is shown in Figure 1, in which the sequence comparison time of the GSP
is compared with that of the unaided host.
Host execution times were measured running a C implementation of the
dynamic programming algorithm. We
deliberately made this program as fast as possible, to avoid biasing the
results in favor of the GSP. Execution
time is shown on the vertical axis, plotted against the number of nucleotides. For these measurements, both sequences are
the same length.
Figure 1
shows an average performance improvement of about 15x. Note that despite the appearance of the
graph, GSP performance scales quadratically.
As expected, using special purpose hardware to solve a problem while
leaving the algorithm unchanged improves performance by a constant factor.
The GSP
board is clocked at 10 MHz, and requires 6 clock ticks = 600ns to calculate
each entry in the dynamic programming array.
The resulting execution times are thus very close to 600(n+1)2/109,
as the time to initialize the system and retrieve the alignment using the back
pointers is negligible.
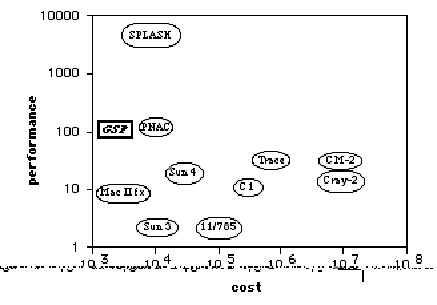
The
cost-performance plane for gene sequence comparison is shown in Figure 2. These numbers are taken from [8] and [7],
with the points for the GSP and the unassisted Mac II added by the authors. Performance/cost ratios for the machines in
Figure 2 are shown in Table 1. In both
Figure 2 and Table 1, performance is taken to be the reciprocal of execution
time. Note that both axes in Figure 2
are logarithmic.
We see that
for gene sequence comparison, the GSP has the most favorable performance/cost
ratio of all machines but Splash. The
performance improvement of Splash is quite significant, due its use of a
systolic array architecture to exploit parallelism. It is unlikely that the GSP could achieve
comparable performance without a significant increase in cost. Unlike Splash, however, the GSP is
alignment-preserving, and while it appears that Splash can be modified to
produce alignments [19], no performance results have been reported as yet. The GSP can also be programmed by a novice,
using the Macintosh user interface. Finally, the GSP project was completed in a single
man-year, with the actual design and debugging of the board requiring only
three months. This suggests that future
versions, for which more time and resources will be available, can achieve even
better results.
7.0 Conclusions and Future Work
The initial goal of this project
was to construct an attached processor for gene sequence analysis that would
offer an order of magnitude improvement in performance over personal computers
without any increase in cost. Our
efforts were successful: the Gene Sequence Processor has met or exceeded design
specifications. We were also able to
advance our work in rapid prototyping, in particular with regard to FPGA's and
PCB fabrication, using the environment of the Rapid Prototyping Facility.
We believe
our emphasis on building working hardware has paid off. Results obtained from functioning machines
are inherently more credible than simulation studies, and are more likely to be
reproduced elsewhere. Our efforts in
this area indicate that technological advances are making the production of
working hardware feasible in environments where formerly simulation was all
that could be attempted. We expect this
trend to continue, bringing with it an increased rigor and credibility to
computer architecture as an intellectual discipline.
Much remains
to be done with the Gene Sequence Processor.
Our next goal is to add another order of magnitude of performance to GSP
without a corresponding increase in cost.
We hope to achieve this in several ways.
The current GSP exploits none of the parallelism inherent in dynamic
programming algorithms, a significant source of performance for Splash. Future implementations of GSP will attempt to
utilize more parallelism, within existing cost constraints. Other possibilities include adding an SRAM
cache to exploit the highly regular memory reference pattern of the dynamic
programming algorithm, using a faster clock, and incorporating pipelining into
the design.
8.0 Acknowledgements
The authors gratefully
acknowledge the assistance of the Whitaker Foundation, whose generosity made
the construction of the GSP possible.
Support for the Rapid Prototyping
Facility was provided by a variety of sources, including Actel, Altera, Xilinx,
Sun Microsystems, Viewlogic, National Semiconductor, and Direct Imaging
Incorporated. Additional support was
provided by the Thayer School of Engineering, and the National Science
Foundation, award #CDA-8921062.
The first
author can be reached electronically
at barry.fagin@dartmouth.edu.
9.0 References
[1] Computer
Applications in the Biological Sciences,
IRL press, Oxford, UK.
[2] Bishop, M.J. and Rawlings, C.J., Nucleic Acid and Protein Sequence
Analysis, IRL Press, Washington D.C.
1987.
[3] Collins, J.F. and Coulson, A.F.W.,
"Applications of Parallel Processing Algorithms for DNA Sequence
Analysis", Nucleic Acids Research, Vol.
12, No. 1, 1984, pp 181-192.
[4] Mukherjee, A., "Hardware Algorithms for Determining
Similarity Between Two Strings". IEEE
Transactions on Computers, April
1989, pp 600-603.
[5] Gotoh, Osamu, and Tagashira,
Yusaku, "Sequence Search on a Supercomputer", Nucleic Acids Research, Vol.
14, No. 1, 1986, pp 57-64.
[6] Core, Nolan et. al., "Supercomputers and
Biological Sequence Comparison Algorithms", Computers and Biomedical Research,
Vol. 22, 1989, pp 497-515.
[7] Lopresti, Daniel, "Sequence Comparison on Commercial
Supercomputers", Supercomputing Research Center Technical Report SRC-TR-89-010,
October 1989.
[8] Gokhale, Maya et. al., "Building and
Using a Highly Parallel Programmable Logic Array", Computer, Vol. 24, No. 1,
January 1991, pp 81-89.
[9] Lopresti, Daniel, "P-NAC: A Systolic Array For Comparing
Nucleic Acid Sequences", Computer,
Vol. 20, No. 7, July 1987, pp 98-99.
[10] Fagin, Barry
"Quantitative Measurements of FPGA Utility in Special and General Purpose
Microprocessors", Journal of VLSI
Signal Processing, Special issue on
Field Programmable Gate Arrays (to appear).
[11] Erickson, Adam and Fagin,
Barry, "Calculating the FHT in
Hardware", IEEE Transactions on
Signal Processing, June 1992., pp
1341-1353.
[12] "Using Antifuse-Based
FPGAs in Performance-Critical Digital Designs", Proceedings of the 4th Microelectronics
System Education Conference and Exposition, San Jose, CA, 1991.
[13] Kruskal, Joseph B., An Overview of Sequence Comparison: Time
Warps, String Edits, and Macromolecules,
SIAM Review, April 1983, Vol 25, No 2, pp 201-237.
[14] Sellers, Peter H., On the Theory and Computation of
Evolutionary Distances, SIAM Journal of Applied Mathematics, June 1974,
Volume 26, No. 4, pp 787-793.
[15] Xilinx Programmable Gate Array Databook,
Xilinx Incorporated, 1990.
16] El Gamal, Abbas "An Architecture for Electrically
Configurable Gate Arrays", IEEE Journal of Solid State Circuits, Vol. 24, No. 2, April 1989.
[17] El-Ayat, Khaled et. al., "A CMOS
Electrically Configurable Gate Array",
IEEE Journal of Solid State
Circuits, Vol. 24, No. 2, April
1989.
[18] "Using Reprogrammable
Gate Arrays in Performance-Critical Digital Designs", Proceedings of the
3rd Microelectronics System Education Conference and Exposition, San Jose, CA, 1990.
[19] Hoang, Dzung, Brown University Department of
Computer Science Technical Report, in preparation.